- 統合医療機能性食品国際学会
- お知らせ
- 【寄稿】非コード性内因性アンチセンスRNAによる自...
トピックス
【寄稿】非コード性内因性アンチセンスRNAによる自然免疫応答遺伝子の発現制御
- トピックス
- 2021.12.15

ICNIM2018にてAHCC®の乳癌細胞における抗腫瘍作用メカニズムに関する研究で優秀研究報告賞を受賞した立命館大学薬学部・木村富紀特任教授より、ご寄稿頂いたので以下に掲載する。
はじめに
近年に至るまで、RNAはゲノムDNAがコードするタンパク質情報を細胞質において発現するためのblueprintと位置付けられていた[1]。しかしながら、ヒトゲノムプロジェクトに引き続くENCODEプロジェクトの結果、ゲノムにおける遺伝子領域はわずか3%足らずに過ぎないにも関わらず、全ゲノム領域の75%相当が転写されている事が明らかにされた[2, 3]。更には、タンパク質情報をコードしない非コード性RNA (protein non-coding RNAまたはncRNA)の多くには遺伝子の発現を制御する制御性RNAとしての機能が賦与されている事が明らかになり[4]、タンパク質を発現するためのblueprintと見なされていたRNAの概念を根底から崩すことになった。
ncRNAによる遺伝子発現制御の概要
これらncRNA、中でも200塩基長以上のlong ncRNA (lncRNA)が制御性RNAとして示す遺伝子発現調節作用には、エピジェネティック制御から転写制御、転写後性制御、更には翻訳並びに翻訳後性制御に至るまで多岐にわたる事が知られており、これはlncRNAがシャペロンとしてそれぞれの制御効果に係わるRNA結合タンパク質を標的分子にもたらす結果と考えられている[5]。
非コード性内在性アンチセンスRNA (Natural Antisense TranscriptsまたはNAT)によるインターフェロン-alpha1遺伝子(IFNA1)mRNA (Interferon-alpha1 mRNAまたはIFN-α1 mRNA)の転写後性発現調節
私の研究室では、このlncRNAの中でもNATを例にとり、標的mRNAの転写後性発現制御効果の分子メカニズムと、その機能ドメインが生体において果たす有意な生物効果に着目し、研究を進めてきた[6]。その結果、ウイルス感染に対する宿主自然免疫応答の主たるエフェクターをコードするIFN-α1 mRNAの発現にあたっては、IFNA1遺伝子の逆鎖から転写されるIFN-α1 antisense RNA (IFN-α1 AS)が転写後性にmRNAの安定性を制御することを見出している[7, 8]。
このIFN-α1 ASによるIFN-α1 mRNAの安定性の制御には、以下に述べる二つのメカニズムが係っていた。
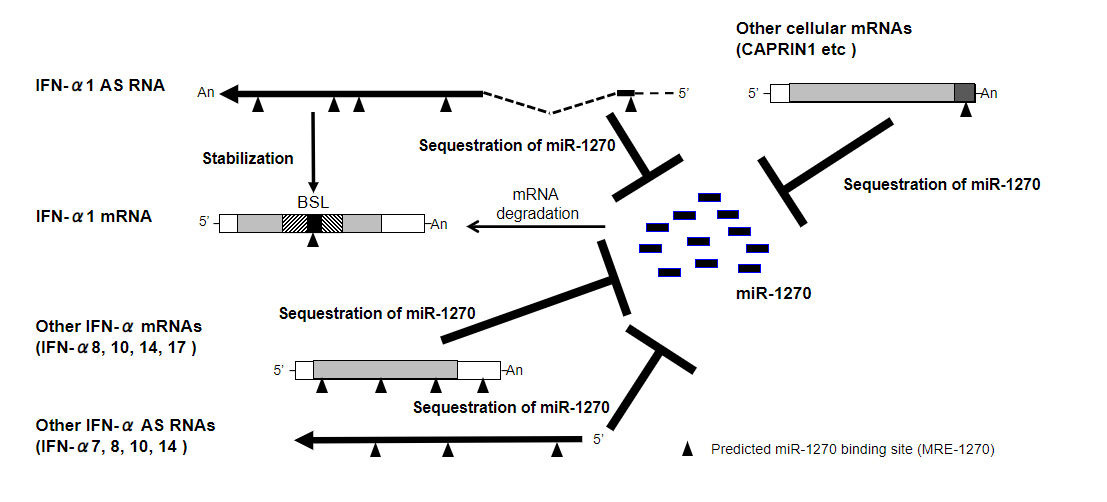
- 第一には、IFN-α1 mRNAが形成するRNA二次構造中のconserved secondary structure (Fig.1、IFN-α1 mRNA斜線部)に存在するbulged-stem loop (BSL)をIFN-α1 ASが直接認識し、作用点局所においてA フォーム二本鎖が形成される事によった[7, 9]。このAフォームRNA二本鎖が形成されると、minor groove領域の開大化を伴う局所コンフォメーションが変化し、mRNA安定化タンパク質として知られるthe embryonic lethal-abnormal visual protein (HuR)の結合増大や、逆にmRNAの不安定化をもたらすtristetraprolin (TTP)やKH-type splicing regulatory protein (KSRP)の解離が促されるものと考えられた。
- 第二には、IFN-α1 mRNAを認識しその分解をもたらすmicroRNA (miR)-1270に対し、IFN-α1 ASがcompeting endogenous RNA (ceRNA)として機能し、miR-1270の作用を阻害する事によった。先に述べたIFN-α1 ASが認識するIFN-α1 mRNA上のBSLとその近傍領域には、miR-1270が結合するmicroRNA-response-element (MRE)-1270が存在する。ところが、IFN-α1 ASもMRE-1270を共有するため、本来ならばIFN-α1 mRNAに結合するべきmiR-1270をIFN-α1 ASが吸着し、結果としてmiR-1270による分解誘導を免れたIFN-α1 mRNAはその発現が安定化した (Fig.1、IFN-α1 AS、Sequestration of miR-1270; 矢頭はMRE-1270を示す)[8]。
-α8、-α10、-α14 AS並びにCAPRIN1 mRNAを始めとするmRNA群にも共有されている事が明らかとなった。すなわち、IFN-α1 ASはこれらのRNAとceRNAネットワークを形成し、共有するMRE-1270を介してmiR-1270が示す陰性変力作用に拮抗し、IFN-α1 mRNAの安定化を通じてその発現を転写後性に増大すると見なされた (Fig.1)[8]。
創薬シーズとしてのIFN-α1 AS機能ドメイン
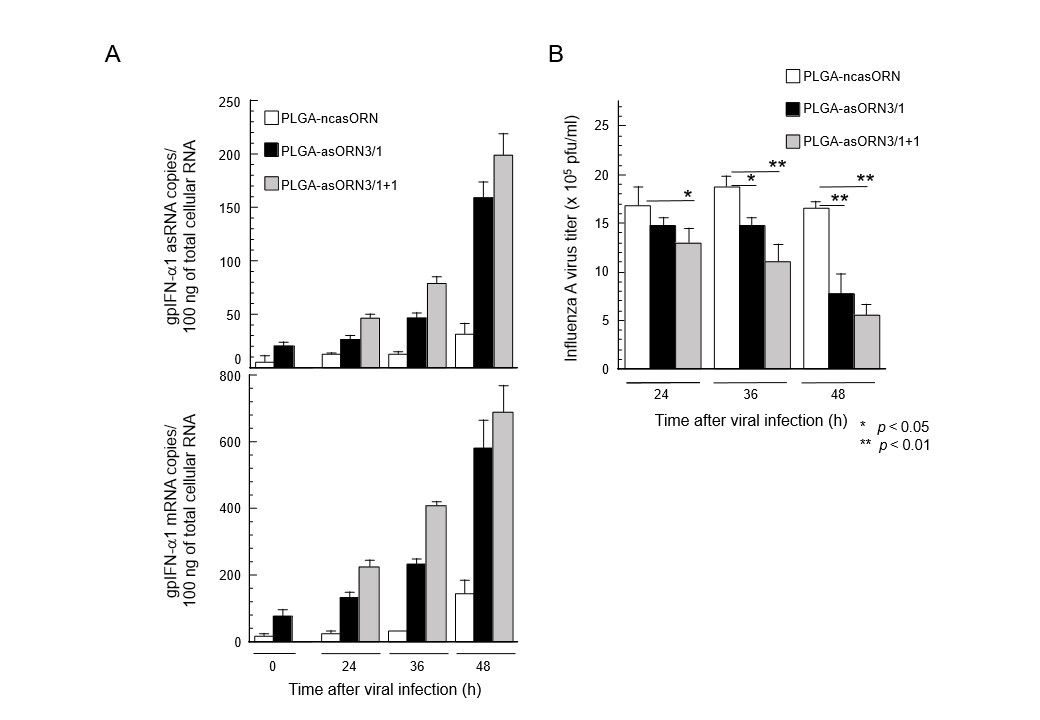
上述したメカニズムによりIFN-α1 mRNAの発現を安定化するIFN-α1 AS上の二種類の機能ドメインを用いて、抗ウイルスヒト自然免疫応答の調節を可能にする創薬の可能性を検討した。この目的で、これら二つの機能ドメインの塩基配列からなるantisense oligoribonucleotide (ORN)を合成し、A型インフルエンザウイルスの気道感染により作製したモルモット感染モデルに投与し、その抗ウイルス効果を比較検討した。モルモット胎児線維芽細胞を用いた予備実験において、MRE配列ドメインからなるasORN3は、BSLを直接認識する配列と比較して、モルモットIFN-α1 mRNAの発現レベルをより高値に誘導していた(結果省略)。そこで、このasORN3をモルモット感染モデルの気道内に投与したところ、気道組織においてその投与量に比例してmiR-1270の吸着によりモルモットIFN-α1 ASの発現を増大させるとともに (Fig.2A、top)、モルモットIFN-α1 mRNAの発現レベルを上昇させた(Fig.2A、bottom)。このモルモットIFN-α1 mRNAの発現増大は感染局所における抗ウイルス応答を誘導し、asORN3の投与量に比例してより早期にかつ有意にA型インフルエンザウイルス力価を低下させた(Fig.2B)。以上のモルモットモデル動物を用いたウイルス感染実験より、IFN-α1 AS上のmRNA発現制御ドメインは感染局所において有効な抗ウイルス自然免疫応答を誘導し、対象動物と比較しより有意に感染力価の低下をもたらす事が証明された[10, 11]。
おわりに
本稿では、lncRNA、中でもNATを例に取り、標的mRNAの転写後性発現制御効果の分子作用メカニズムと、その機能ドメインが生体において有意な生物効果を誘導することを解説した。NATを始めとするlncRNAが示す遺伝子発現調節効果は多岐にわたり、その発現の変調ががんの発症や心血管疾患を始め、多数の疾患に影響を及ぼすことは今や周知となっている。今後、lncRNA研究が生物学、生理学領域の新たな知見の創出につながり、疾病の治療と予防に貢献すると期待される。
参考文献
F. Crick, Nature, 227 (1970) 561-563.
[2] An integrated encyclopedia of DNA elements in the human genome
Encode Project Consortium, Nature, 489 (2012) 57-74.
[3] Landscape of transcription in human cells
S. Djebali, et al., Nature, 489 (2012) 101-108.
[4] Regulatory RNA
A.K. Eggleston, et al., Nature, 482 (2012) 321.
[5] Regulation of innate immune responses by long noncoding RNAs
N. Akimitsu, SEIKAGAKU, 87 (2015) 385-388.
[6] Non-coding natural antisense RNA: mechanisms of action in the regulation of target gene expression and its clinical implications
T. Kimura, YAKUGAKU ZASSHI, 140 (2020) 680-700.
[7] Stabilization of human interferon-alpha1 mRNA by its antisense RNA
T. Kimura, et al., Cell Mol Life Sci, 70 (2013) 1451-1467.
[8] Interferon-alpha competing endogenous RNA network antagonizes microRNA-1270
T. Kimura, et al., Cell Mol Life Sci, 72 (2015) 2749-2761.
[9] Regulation of inducible gene expression by natural antisense transcripts
M. Nishizawa, et al., Front Biosci (Landmark Ed), 17 (2012) 938-958.
[10] A guinea pig IFNA1 gene with antiviral activity against human influenza virus infection
S. Jiang, Front Biosci (Landmark Ed), 24 (2019) 790-797.
[11] IFN-Alpha1 antisense RNA represses human influenza A virus growth in a guinea pig system
R. Sakamoto, et al., Front Biosci (Landmark Ed), 24 (2019) 798-818.

