- ICNIM|AHCC Research Association
- TOPICS
- 【Contributed Article】Non-coding endogenous antisense RNAs...
TOPICS
【Contributed Article】Non-coding endogenous antisense RNAs control the expression of genes involved in innate immune responses
- TOPICS
- 2021.12.15

We are pleased to post the following article contributed by Dr. Tominori Kimura, Professor, Department of Pharmacy, College of Pharmaceutical Sciences, Ritsumeikan University, who won the Best Research Award at the 28th annual meeting of ICNIM (ICNIM2018) for his study on molecular mechanisms of actions of AHCC® on human mammary carcinoma cell growth and migration.
Introduction
Until recently, RNA was thought to only be the blueprint for the cytoplasmic expression of protein-coding sequences encoded by genomic DNA [1]. However, the ENCODE (Encyclopedia of DNA Elements) project, which followed the Human Genome Project, revealed that three-quarters of all genomic regions were capable of being transcribed, of which only 3% were transcribed into protein-coding mRNAs [2, 3]. Furthermore, it has become clear that many protein non-coding RNAs (ncRNAs) are endowed with regulatory activities that control gene expression [4], thus disrupting the notion that RNAs are solely blueprints for protein expression.
Outline of how ncRNAs control gene expression
These ncRNAs, particularly long ncRNAs (lncRNAs) over 200 nucleotides in length, play many crucial roles in the control of gene expression, including at the epigenetic, transcriptional, post-transcriptional, translational, and post-translational levels. lncRNAs are thought to act as chaperones for the association of regulatory RNA-binding proteins with their target molecules[5].
Post-transcriptional regulation of interferon-alpha1 (IFN-α1) mRNA by a non-coding endogenous antisense RNA (also known as a natural antisense transcript, or NAT)
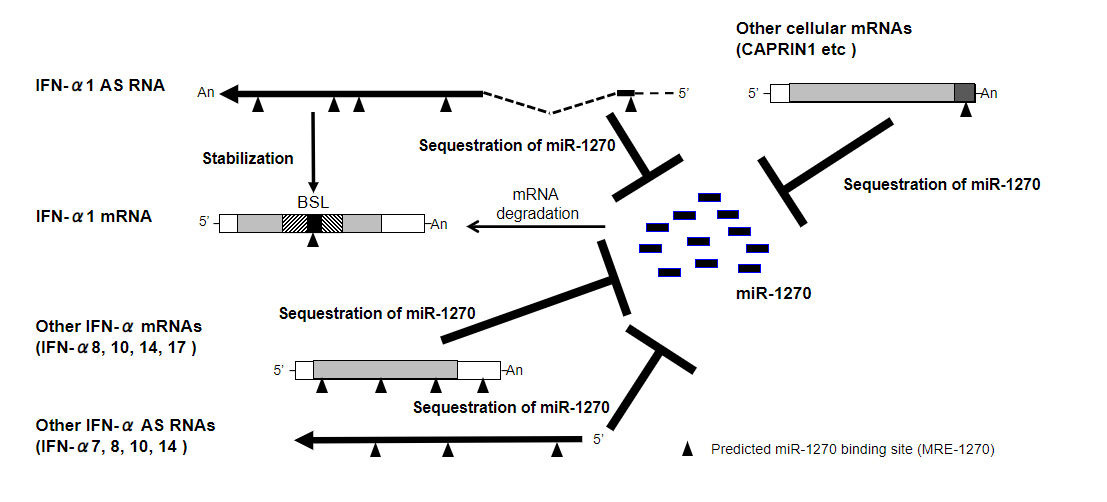
lncRNAs comprise multiple classes of RNA transcripts that regulate expression of protein-coding genes in the mammalian genome, as described above. One such class, natural antisense transcripts (NATs), are transcribed from the opposite strand of a protein-coding or non-protein-coding (sense) gene. We previously identified and characterized one such sense–antisense pair, IFN-α1 mRNA and its antisense partner IFN-α1 AS. We demonstrated that this non-coding NAT plays a critical role in the post-transcriptional regulation of the IFN-α1 gene (IFNA1), and subsequently, IFN-α1 protein production [6]. A ~4 kb, spliced IFN-α1 AS targets a single-stranded region, the bulged-stem loop (BSL), within a conserved secondary structure element of the IFN-α1 mRNA-coding region (Fig. 1, hatched areas of the IFN-α1 mRNA), not in the usual 3′-untranslated region (3′-UTR). Upon recognition of the BSL by IFN-α1 AS in the cytoplasm, the sense–antisense interaction between the complementary transcripts stabilizes the IFN-α1 mRNA [6].
The RNA duplex formed at the site of action is called an A-form RNA duplex, which locally opens the minor groove regions, promoting increased binding of the embryonic lethal-abnormal visual gene (HuR), which is a known mRNA-stabilizing protein, and the dissociation of tristetraprolin (TTP) and KH-type splicing regulatory protein (KSRP), both of which lead to mRNA destabilization [7, 8].
IFN-α1 AS also functions as a competing endogenous RNA (ceRNA) against microRNA (miR)-1270, which recognizes IFN-α1 mRNA, providing an additional, synergistic role for IFN-α1 AS in the post-transcriptional regulation of human IFN-α1 mRNA stability. Overexpression of miR-1270 and transfection of antimiR-1270, complementary to the miR-1270 seed region, revealed that IFN-α1 AS is likely to share miR-1270 response elements (MRE-1270s) with IFN-α1 mRNA. Interactions between IFN-α1 AS and miR-1270 through the response elements would, therefore, titrate miR-1270 away from IFN-α1 mRNA, suggesting that the AS acts as an endogenous decoy, or ceRNA, for IFN-α1 mRNA (Fig. 1, “IFN-α1 AS, Sequestration of miR-1270”; arrowheads depict MRE-1270, see also [9]). Indeed, antimiR-1270-dependent silencing of endogenous miR-1270 caused de-repression of IFN-α1 expression levels [9].
Interestingly, while antimiR-1270 transfection resulted in specific de-repression of miR-1270 targets on both IFN-α1 AS and mRNA, its antagonism of miR-1270 also caused a significant increase in expression of AS and mRNA for specific subtypes of the IFNA family as well as five other cellular mRNAs, including CAPRIN1 (Fig. 1; see also [9]). These results suggest that IFN-α1 AS, together with specific IFN-α AS and mRNA subtypes and the five other mRNAs, act as competing molecules in the ceRNA network against miR-1270.
This coordinated regulatory architecture and the cytoplasmic sense–antisense interaction between complementary transcripts suggest that the maintenance of precise physiological type-I IFN levels via post-transcriptional regulatory mechanisms is a vital function of the innate immune system[6, 9].
Investigating the functional domains of IFN-α1 AS as seeds for drug discovery
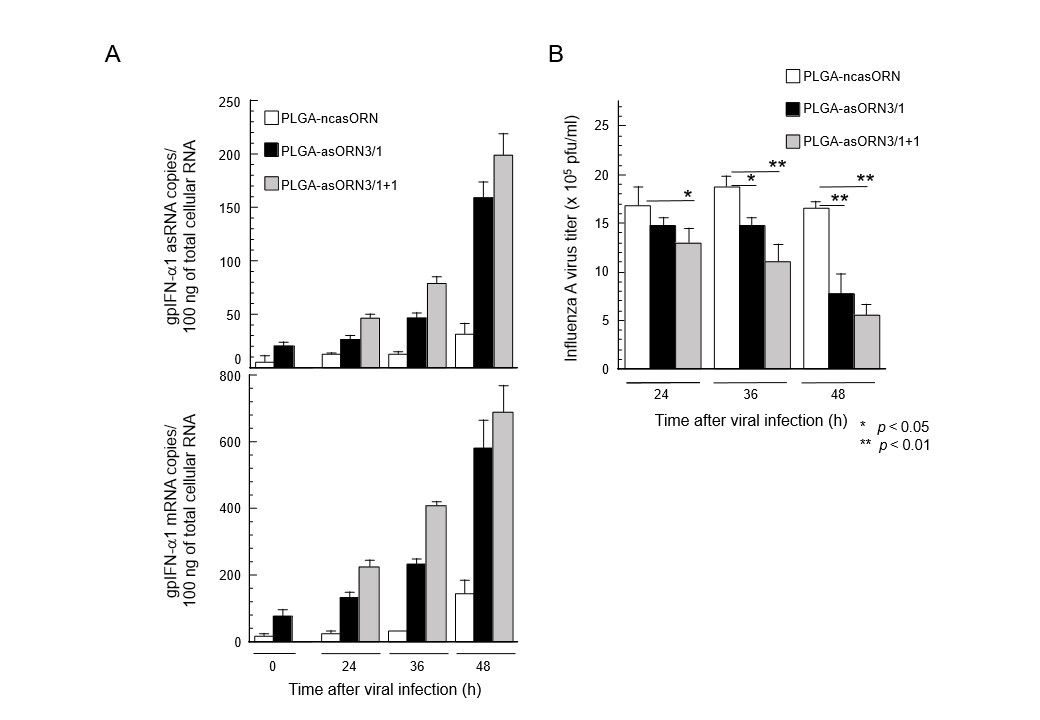
The BSL-recognizing domain and MRE sequence domain of IFN-α1 AS that stabilize IFN-α1 mRNA through the mechanisms described above have been investigated for their potential as novel drugs that modulate the human innate antiviral immune response. To this end, the antiviral efficacy of antisense oligoribonucleotides (asORNs) consisting of the base sequences of either of the functional domains were synthesized and comparatively investigated in a guinea pig infection model involving respiratory tract infection with influenza A virus [10, 11]. In a series of preliminary experiments with guinea pig fetal fibroblasts, an asORN candidate called asORN3, consisting of an MRE sequence domain, induced higher expression levels of guinea pig IFN-α1 mRNA than sequences that directly recognized BSL (results omitted). Therefore, asORN3 was administered into the respiratory tract of the guinea pig infection model. It increased the expression of guinea pig IFN-α1 AS by adsorbing miR-1270 in proportion to the dose in the respiratory tract tissue (Fig. 2A, top) and accordingly raised the expression level of guinea pig IFN-α1 mRNA (Fig. 2A, bottom). The increased expression of guinea pig IFN-α1 mRNA caused an antiviral response at the site of infection, resulting in significant reduction of influenza A virus titers earlier and in proportion to the dose of asORN3 (Fig. 2B) [11].
These viral infection studies in guinea pigs demonstrate that the domain of IFN-α1 AS that controls mRNA expression induces an efficacious innate antiviral immune response at the site of infection that leads to a significantly lower infectivity titer compared with control animals (Fig. 2). We thus succeeded in a proof-of-concept experiment to confirm that the NAT-mRNA regulatory network exerts control over innate immunity in vivo [10, 11].
Conclusions
Our data not only identify mechanisms for post-transcriptional IFNA1 gene regulation but also highlight the role of human antisense transcripts as regulatory RNAs. Growing evidence suggests that antisense transcription plays a key role in a range of human diseases [12]. Activation of the immune system causes the rapid production of IFNs and pro-inflammatory cytokines that orchestrate the developing innate and acquired immune responses to infection. Aberrant or excessive stimulation of these pathways is believed to underlie many inflammatory autoimmune disorders. For example, RNA- and DNA-associated autoantigens in systemic lupus erythematosus have been shown to drive pathological expression of type-I IFN genes and IFN-induced genes through TLR7 and TLR9 activation (reviewed by [13]; see references therein). Our discovery that IFN-α1 AS is a regulator of the IFNA1 gene at the mRNA level raises the possibility that dysfunction of the AS RNA could contribute to autoimmunity through failure to sustain IFNA1 gene expression and function.
In conclusion, IFN-α1 AS adds another level of complexity to the regulation of IFNA1. Further studies of AS RNA may provide a more complete understanding of the regulation of IFNA1 in various physiological processes as well as during autoimmune disease development, and may lead to therapeutic intervention.
Acknowledgment:
I thank Kaley McCluskey, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Reference:
[1] Central dogma of molecular biology
F. Crick, Nature, 227 (1970) 561-563.
[2] An integrated encyclopedia of DNA elements in the human genome
Encode Project Consortium, Nature, 489 (2012) 57-74.
[3] Landscape of transcription in human cells
S. Djebali, et al., Nature, 489 (2012) 101-108.S.
[4] Regulatory RNA
A.K. Eggleston, et al., Nature, 482 (2012) 321.
[5] Regulation of innate immune responses by long noncoding RNAs
N. Akimitsu, SEIKAGAKU, 87 (2015) 385-388.
[6] Stabilization of human interferon-alpha1 mRNA by its antisense RNA
T. Kimura, et al., Cell Mol Life Sci, 70 (2013) 1451-1467.
[7] Regulation of inducible gene expression by natural antisense transcripts
M. Nishizawa, et al., Front Biosci (Landmark Ed), 17 (2012) 938-958.
[8] Non-coding natural antisense RNA: mechanisms of action in the regulation of target gene expression and its clinical implications
T. Kimura, YAKUGAKU ZASSHI, 140 (2020) 680-700.
[9] Interferon-alpha competing endogenous RNA network antagonizes microRNA-1270
T. Kimura, et al., Cell Mol Life Sci, 72 (2015) 2749-2761.
[10] A guinea pig IFNA1 gene with antiviral activity against human influenza virus infection
S. Jiang, Front Biosci (Landmark Ed), 24 (2019) 790-797.
[11] IFN-Alpha1 antisense RNA represses human influenza A virus growth in a guinea pig system
R. Sakamoto, et al., Front Biosci (Landmark Ed), 24 (2019) 798-818.
[12] Long noncoding RNA in genome regulation: prospects and mechanisms
T. Hung, et al., RNA Biol, 7 (2010) 582-585.
[13] Professional type I interferon-producing cells and plasmacytoid dendritic cell precursors
L.Y. Liu., IPC: Annu Rev Immunol, 23 (2005) 275-306.

